|
August 9th 2018: We updated the server to version 1.4.0. (Changelog)
|
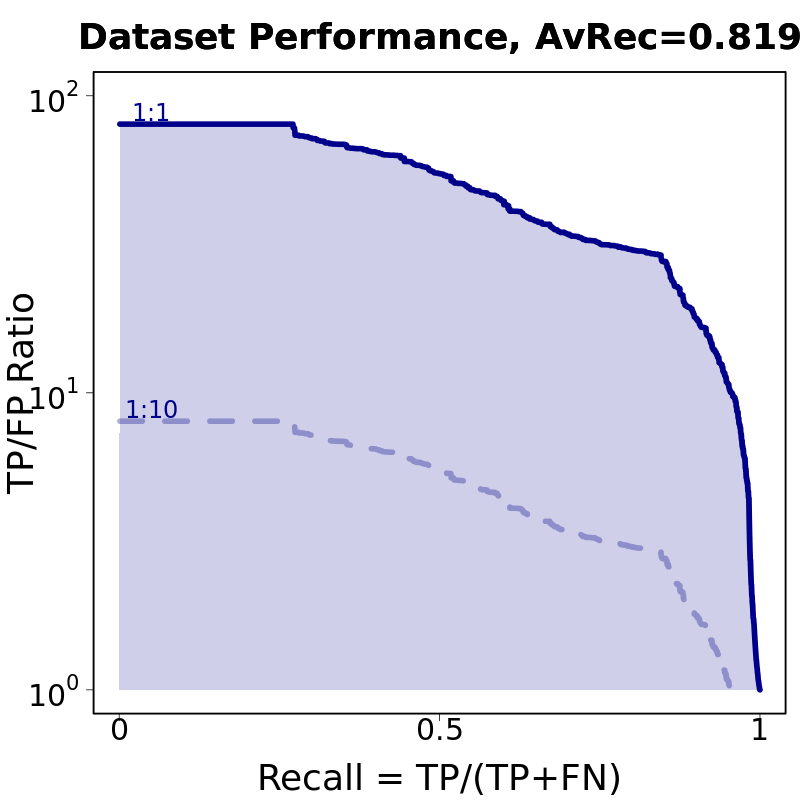
Results for: example run
Input motifs
| # | IUPAC | PWM | reverse Comp. | motif AvRec | fract. occurence | Download |
|---|---|---|---|---|---|---|
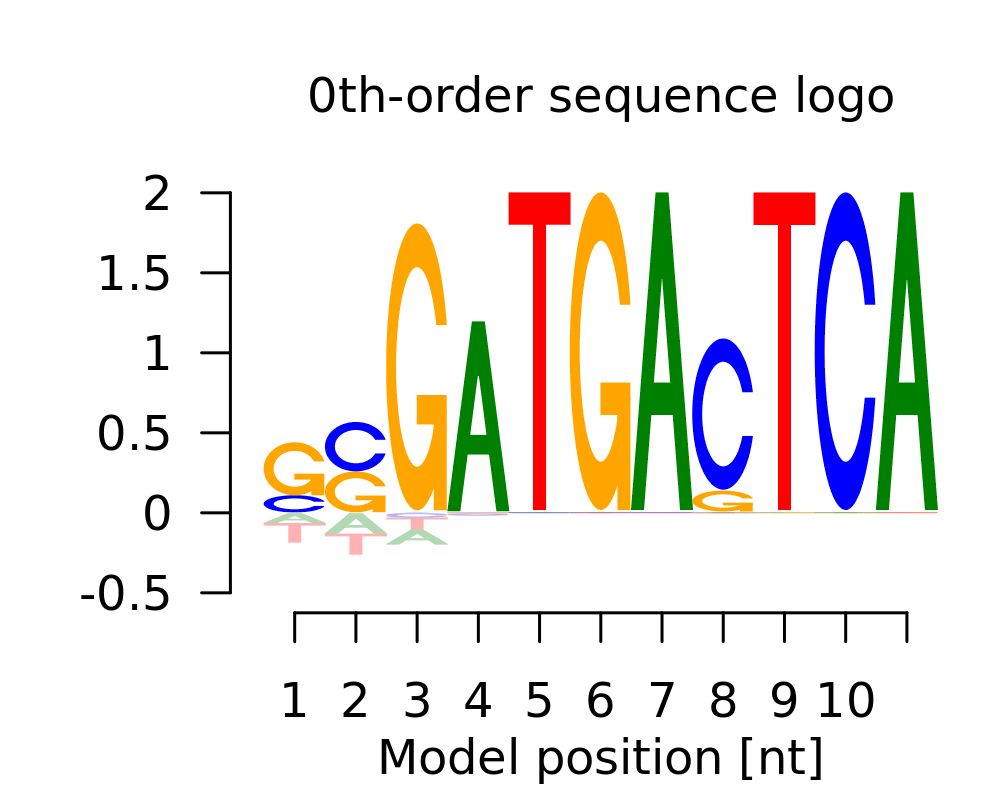
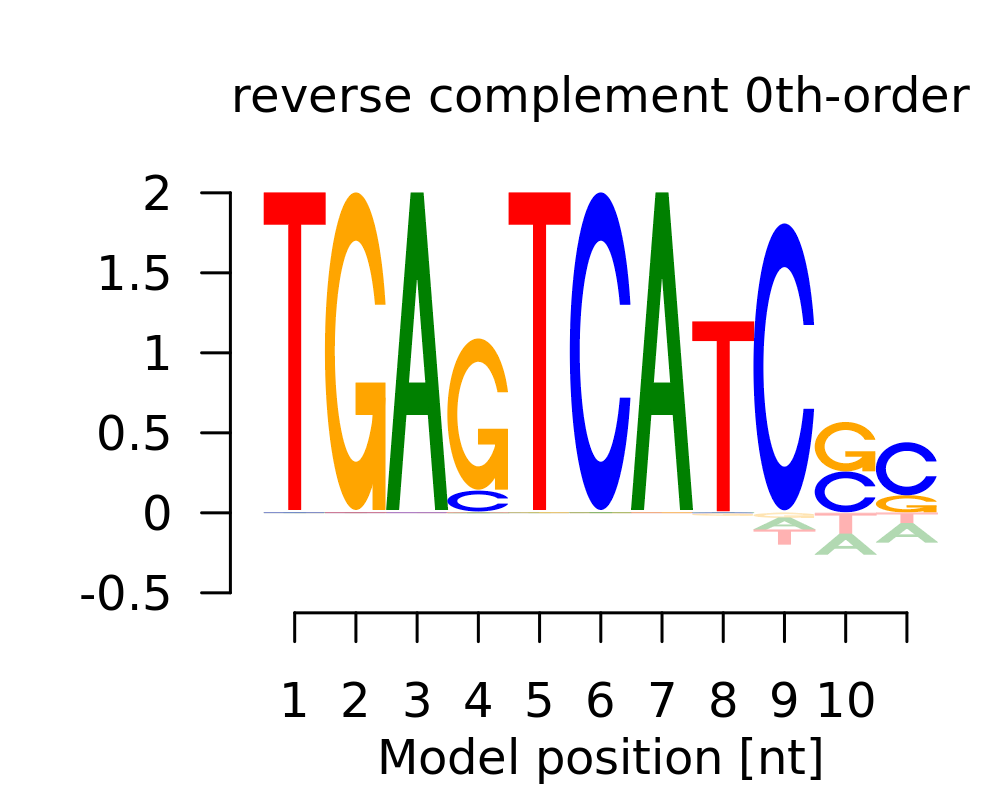
| 1 | SSGATGASTCA |  |
 |
0.839 | 0.977 | |
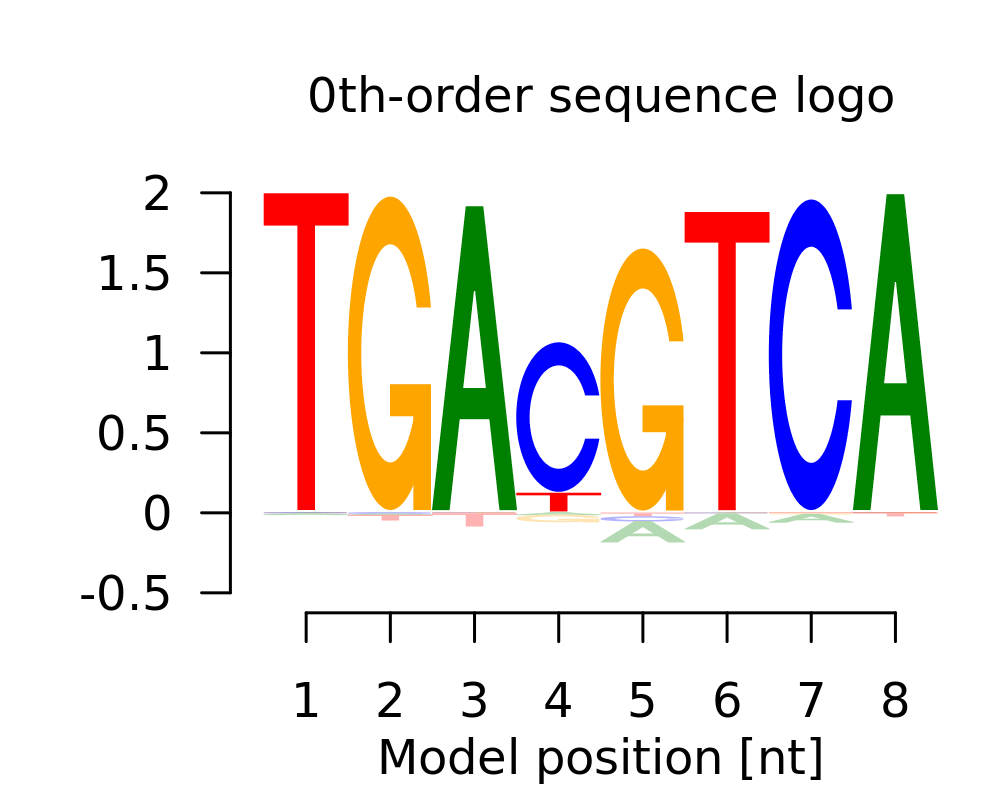
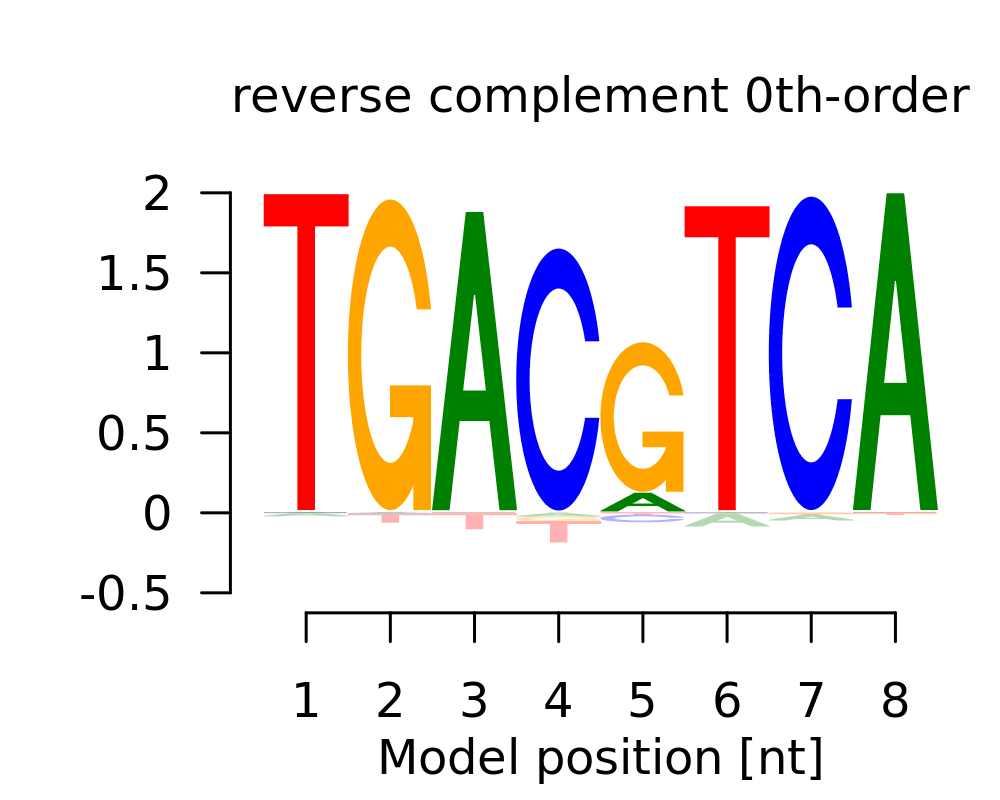
| 2 | TGAYGTCA |  |
 |
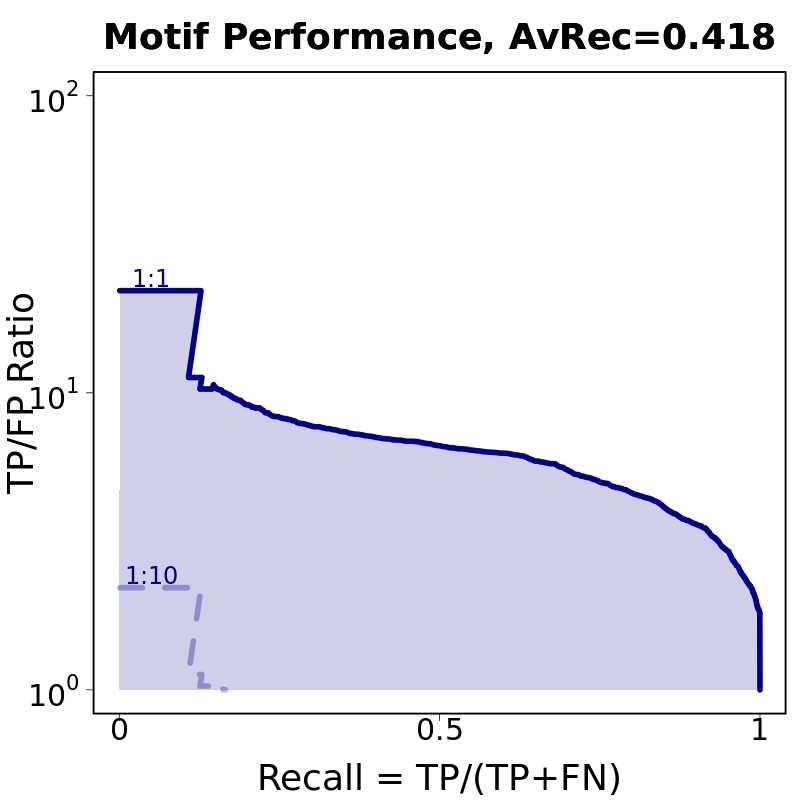
0.418 | 0.938 | |
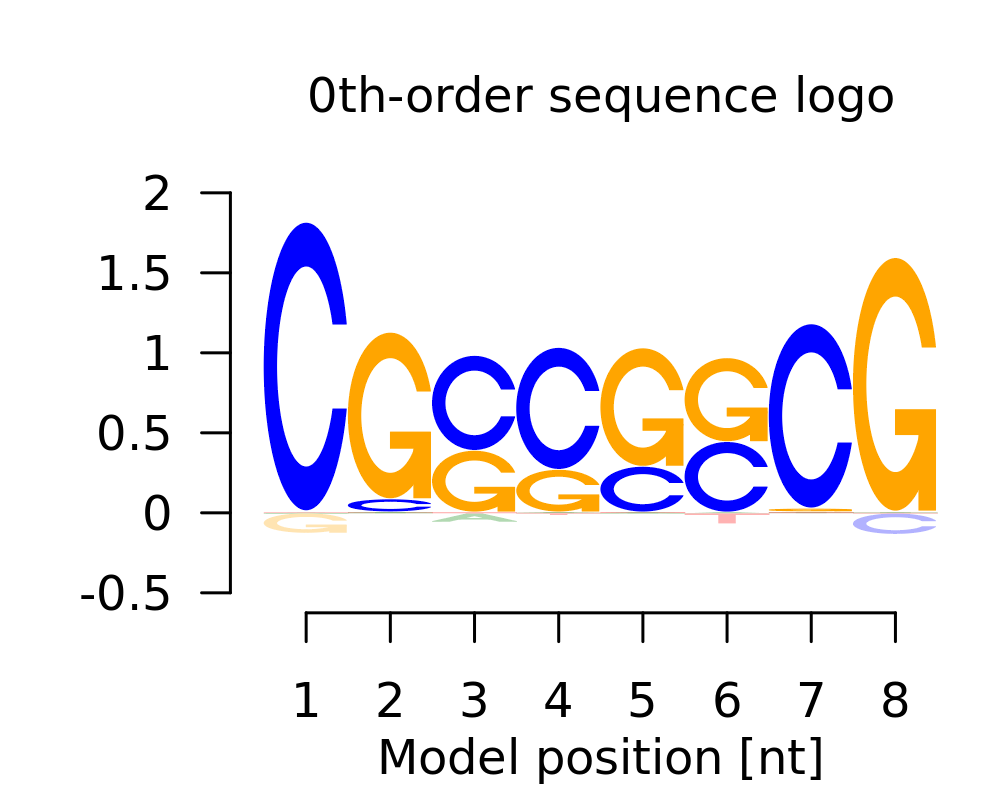
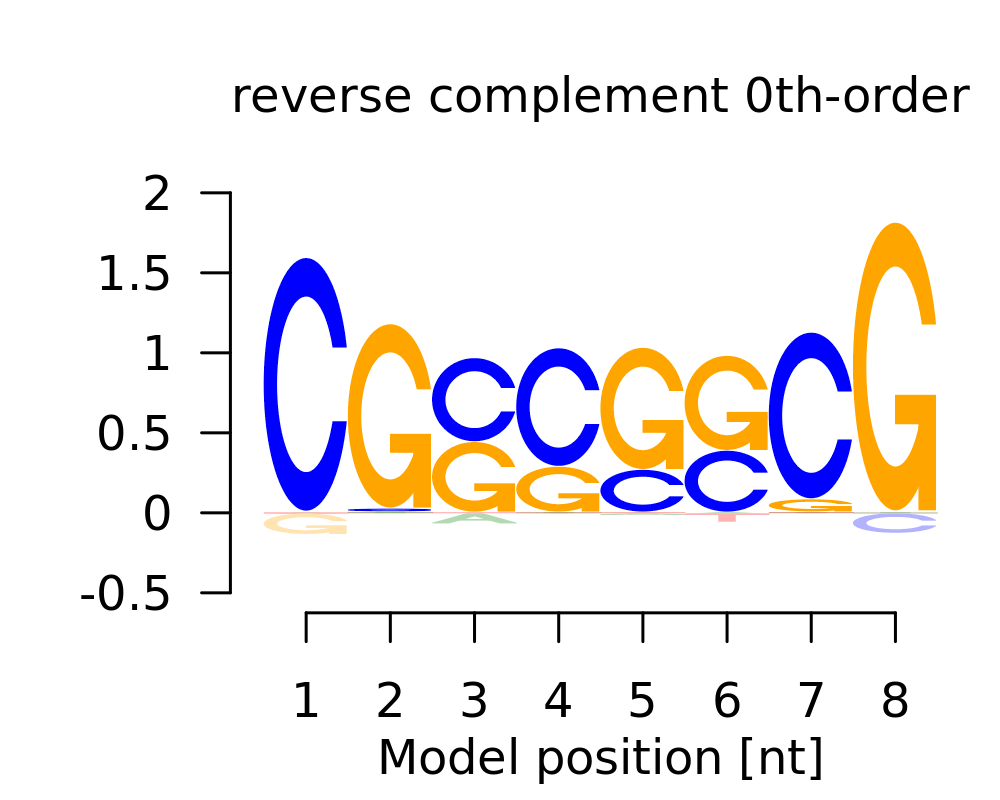
| 3 | CGSSSSCG | 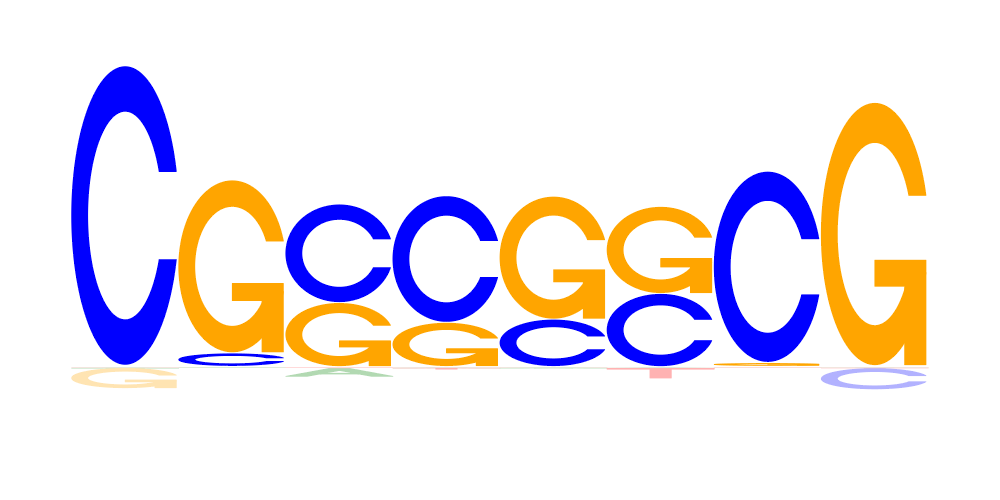 |
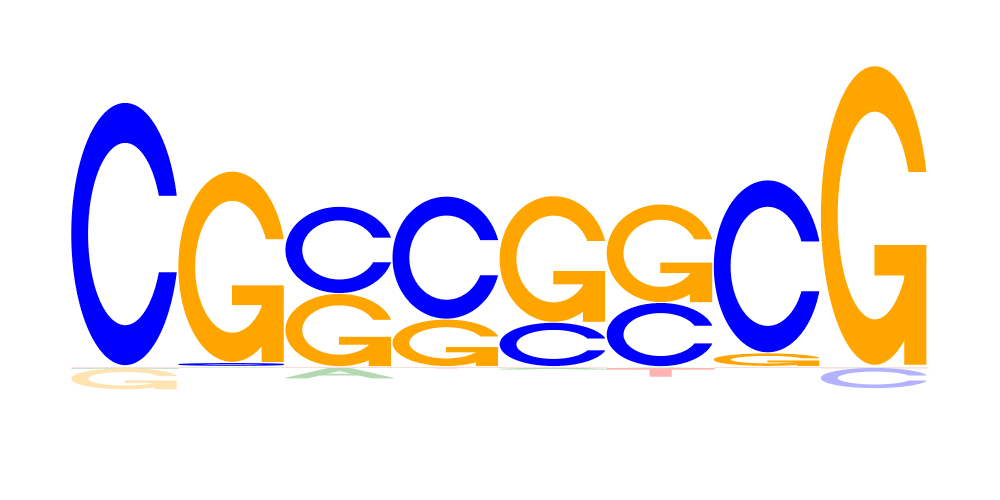 |
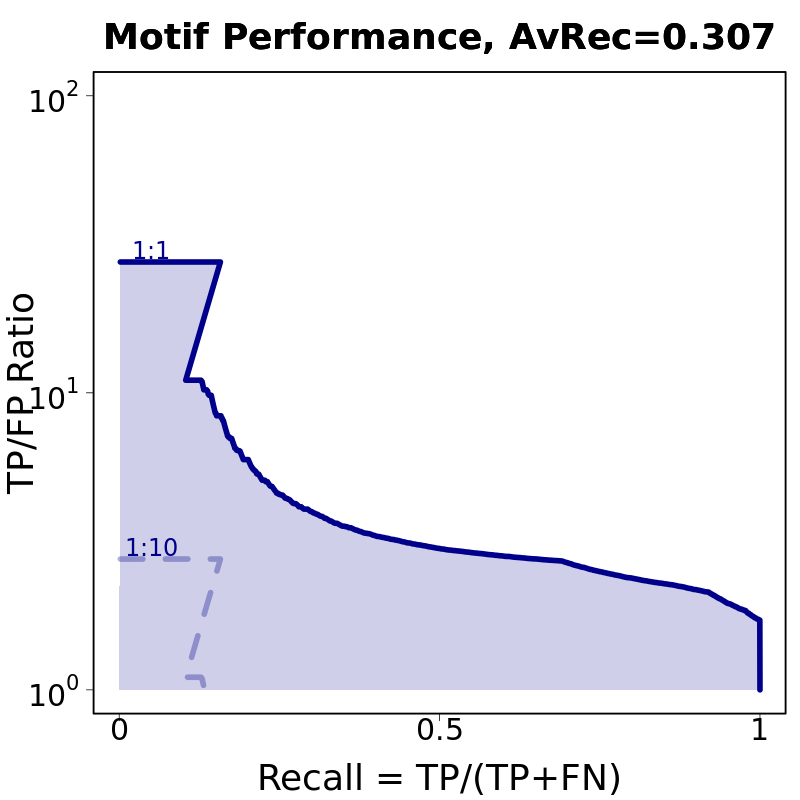
0.307 | 0.366 |
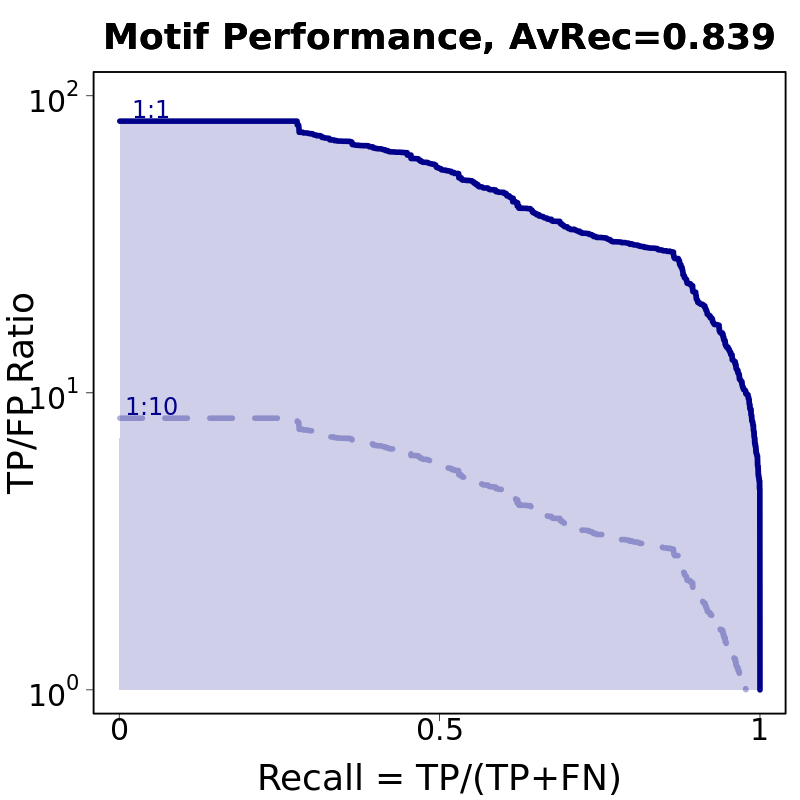
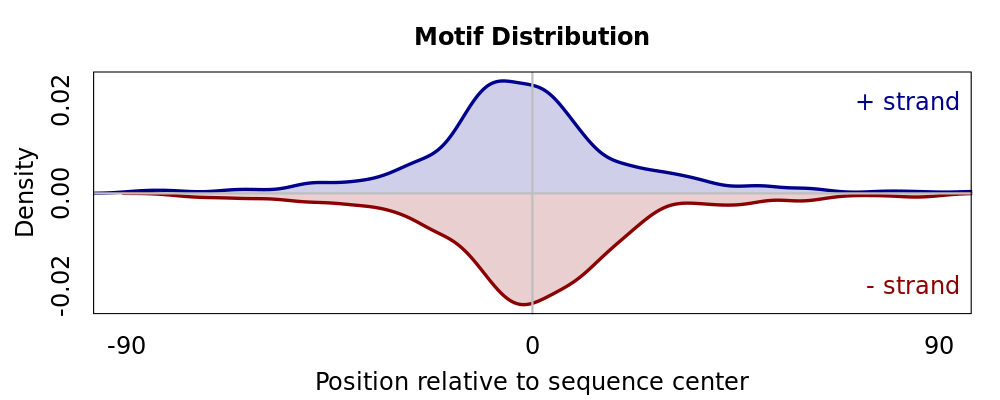
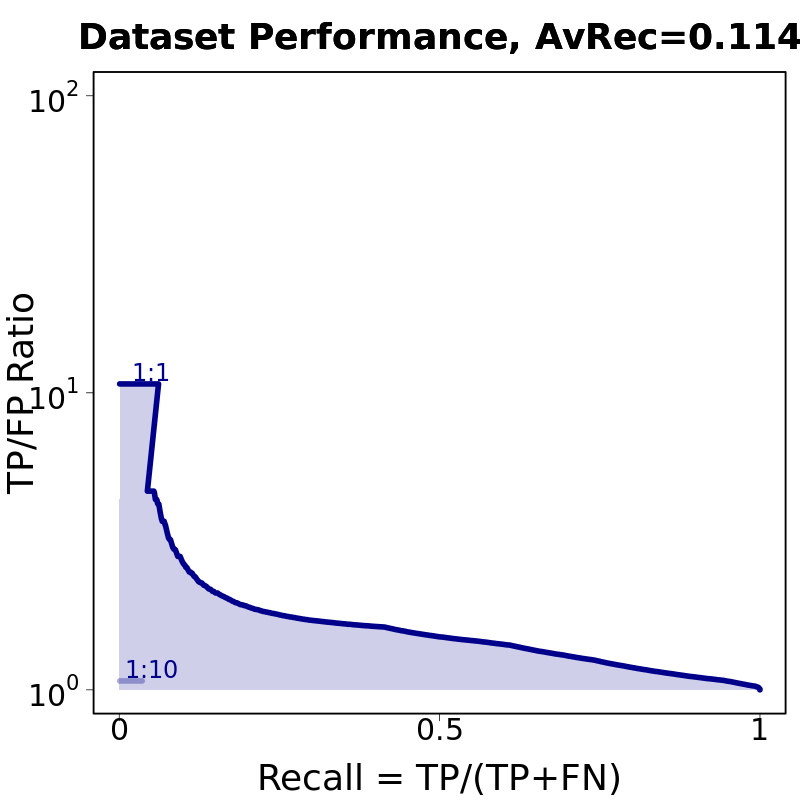
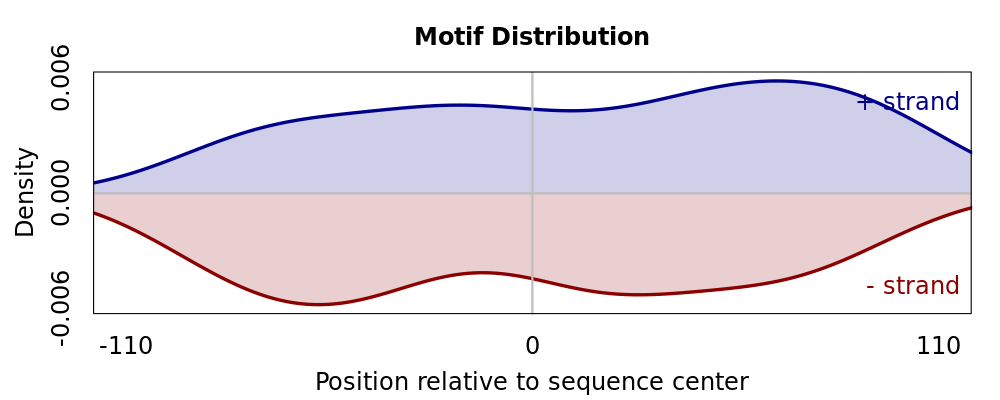
Motif # 3 |
 |
 |
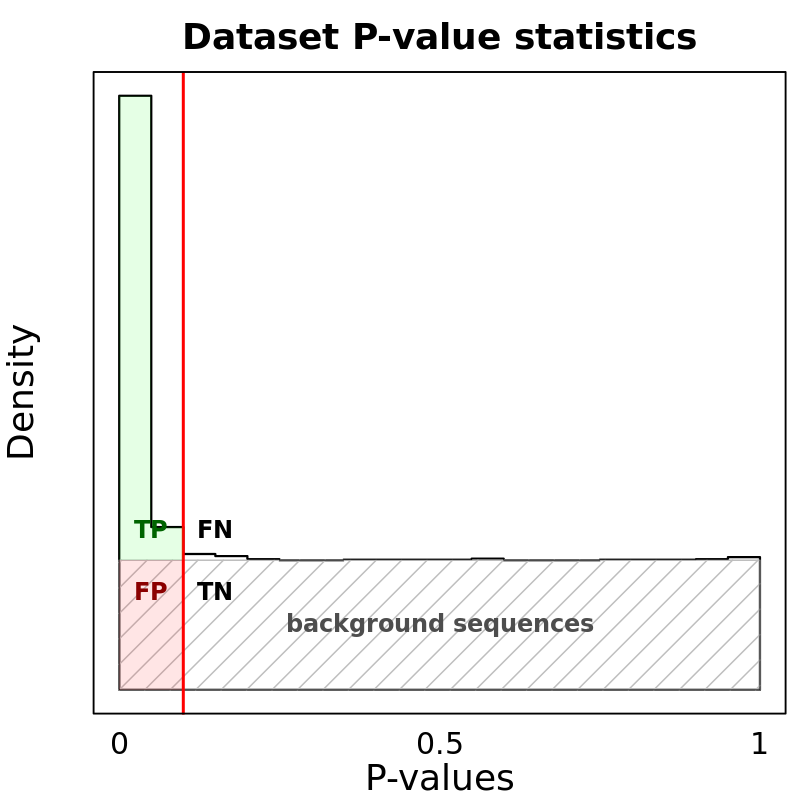
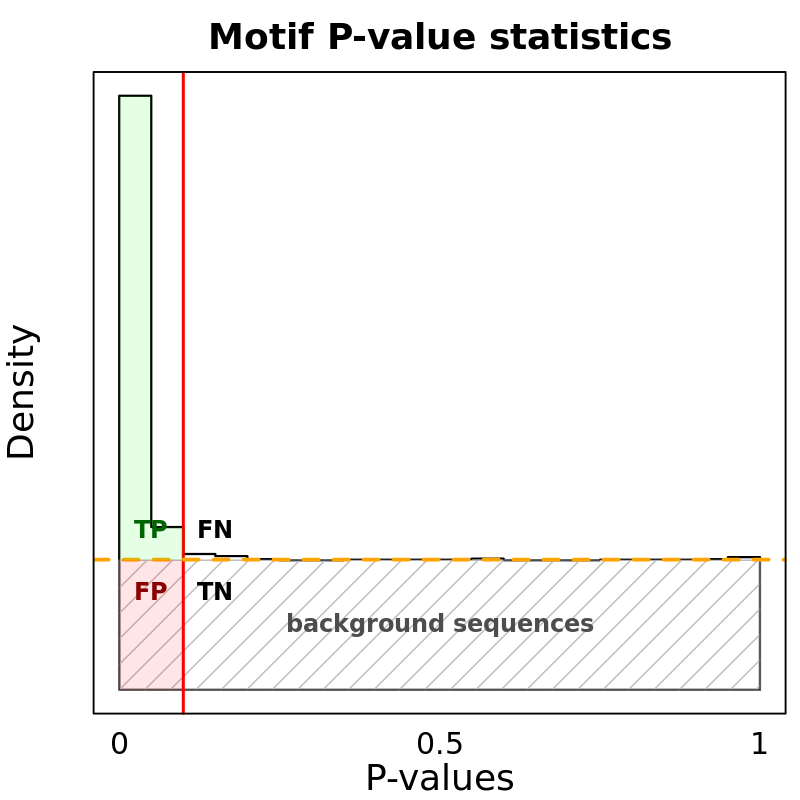
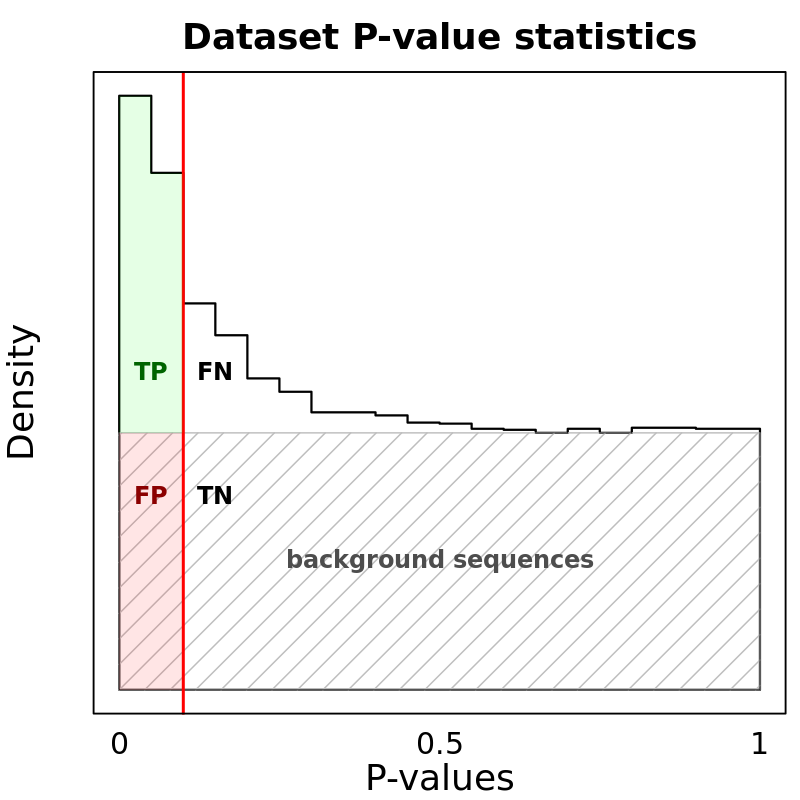
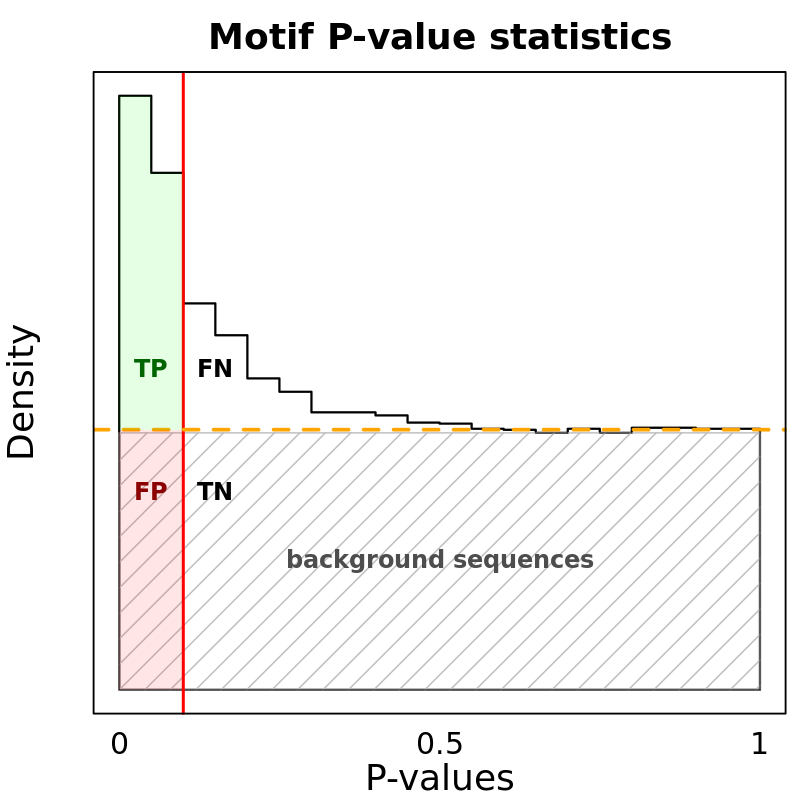
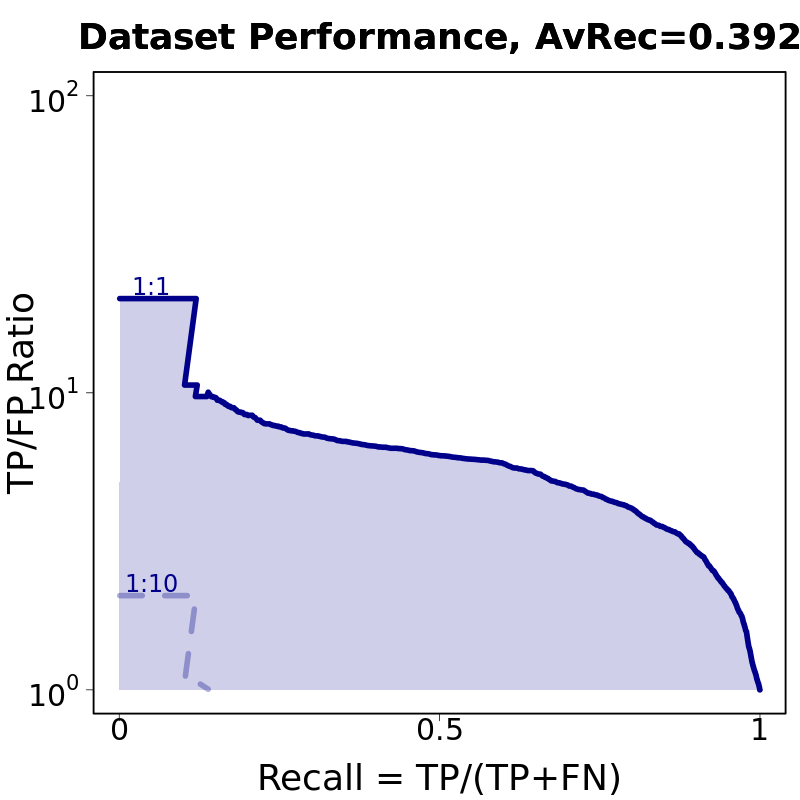
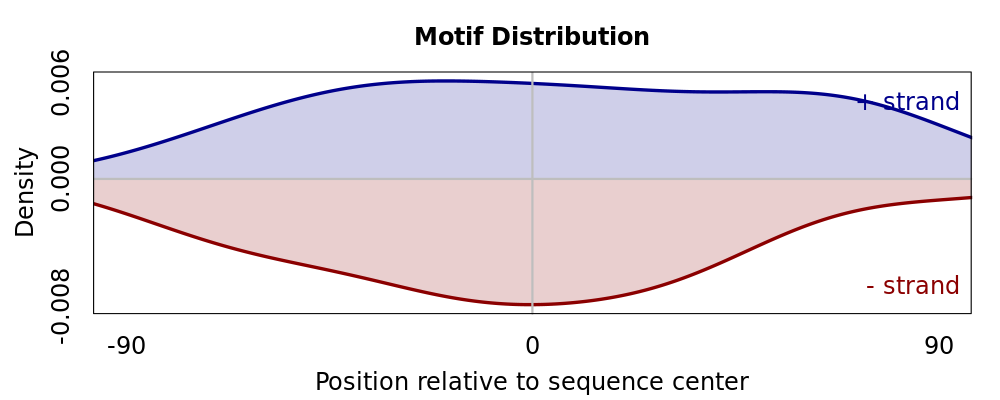
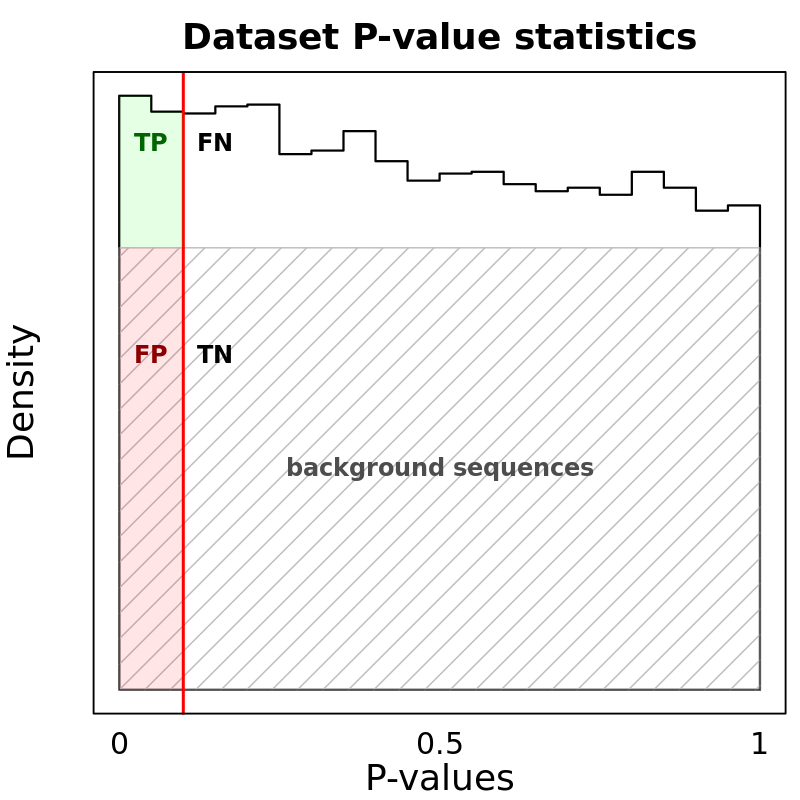
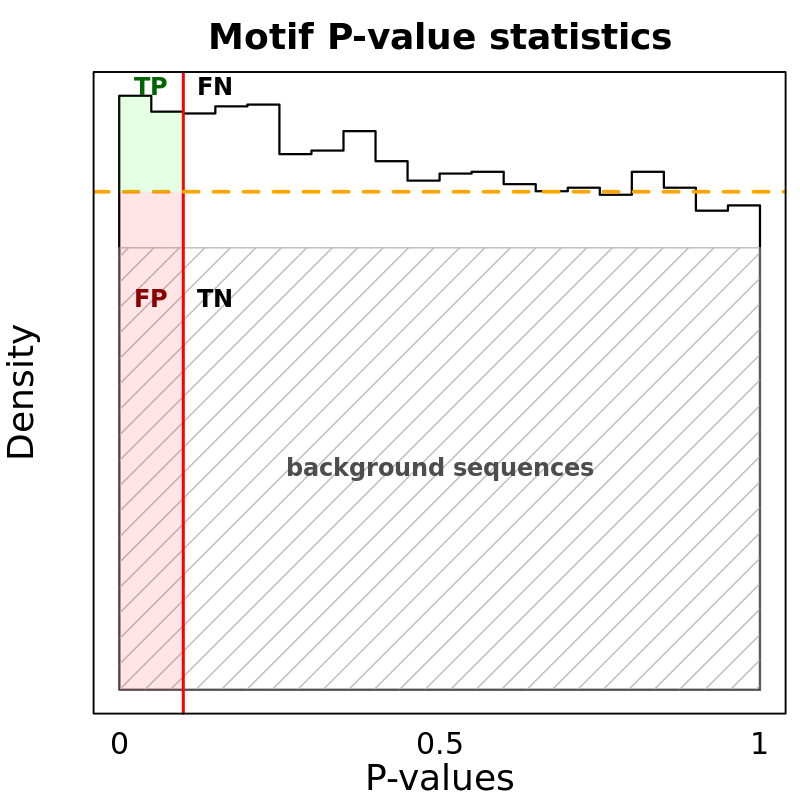
Motif Performance and Motif Distribution on Sequences
|
|
|
|
|
|
Best matches with our motif database
| name | e-value | query motif | database PWM | reverse Comp. | DB entry |
|---|
General settings
| Input Sequences: | ExampleData.fasta |
| Motif File Format: | MEME |
| Motif File: | motif_init.meme |
| Search on both strands: | True |
| Background Model Order: | 2 |
Additional settings
| Run motif scanning: | True |
| Motif scanning p-value cutoff: | 0.0001 |
| Run motif evaluation: | True |
| Run motif-motif compare: | True |
| MMcompare database: | GTRD_v1804_HUMAN |
| MMcompare e-value cutoff: | 0.10 |